![]() What's it?
What's it?
The Omics Discovery Index (OmicsDI) provides dataset discovery across a heterogeneous, distributed group of Transcriptomics, Genomics, Proteomics and Metabolomics data resources spanning eight repositories in three continents and six organisations, including both open and controlled access data resources. The resource provides a short description of every dataset: accession, description, sample/data protocols biological evidences, publication, etc. Based on these metadata, OmicsDI provides extensive search capabilities, as well as identification of related datasets by metadata and data content where possible. In particular, OmicsDI identifies groups of related, multi-omics datasets across repositories by shared identifiers.
![]() How to use it?
How to use it?
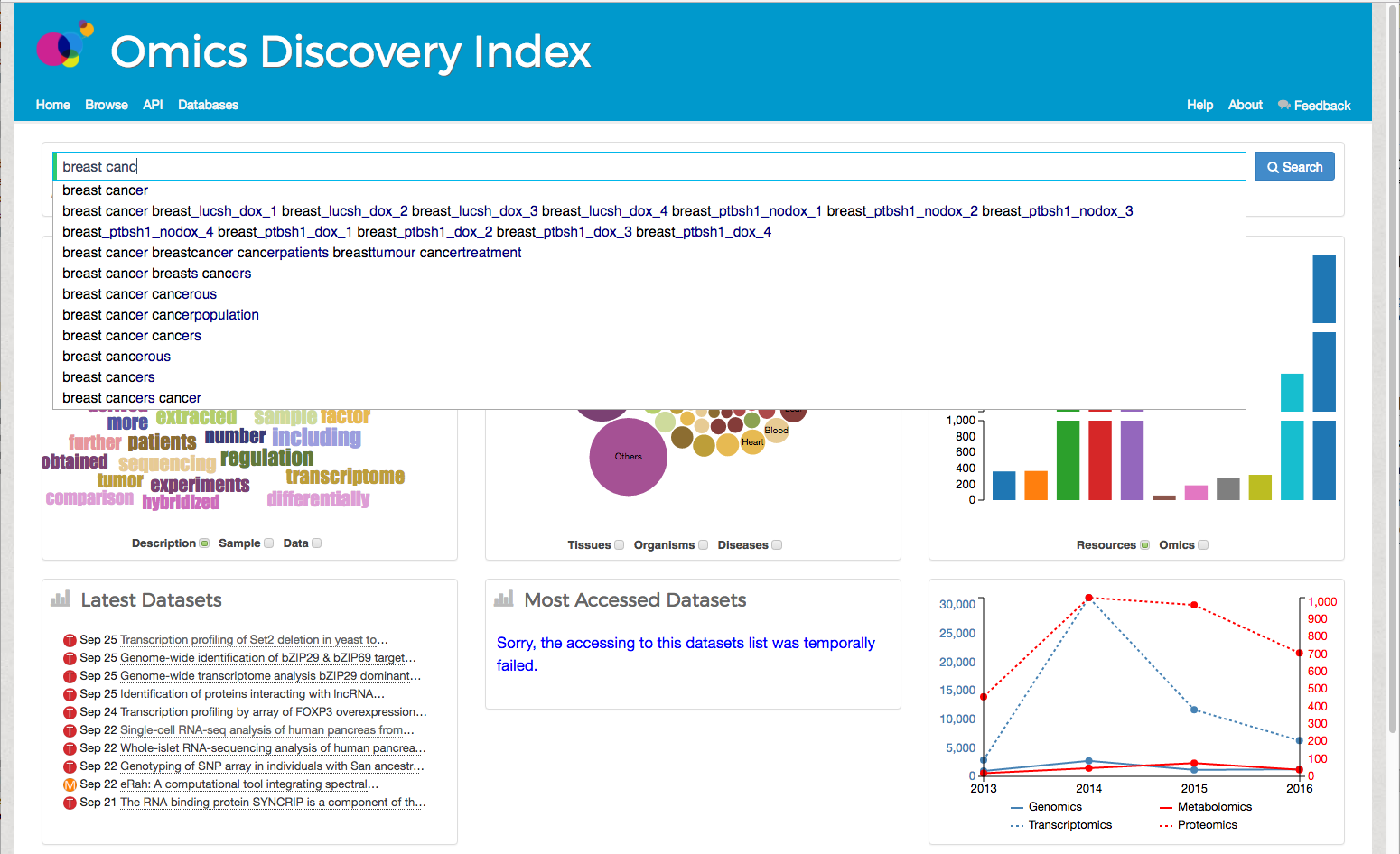
The main goal of the Omics Discovery Index is to provide a platform for searching and linking omics public data. OmicsDI has implemented a unique and novel Search Engine for omics datasets including public and protected data. The OmicsDI Search Box is the main component to searching in OmicsDI. The user can type a set of keywords that will enable the system to find the datasets containing those keywords.If the user uses double quote "breast cancer" in their search the system will try to find the exact sentence in the datasets.The OmicsDI Search Box provides a unique auto-complete feature that enables user to select sentence after typing a subset of keywords. When the user types any text in OmicsDI Search Box, the input is translated into an Apache Lucene query that is then executed to get the search results. The actual query executed is generated following the typical Apache Lucene query syntax in order to provide a generic approach avoiding complex query rearrangements. Multiple search terms separated by white spaces are combined by default in AND logic. Therefore an input text containing for example glutathione transferase is treated as glutathione AND transferase and only entries having both terms will be found. The default order of results is based on their relevance, i.e. the proximity of the terms in the entries. The following characters within queries require to be escaped (using a ‘ \ ‘ before the character to escape) in order to be correctly interpreted: + - & | ! ( ) { } [ ] ^ " ~ * ? : \ / Since Apache Lucene supports regular expression searches (matching a pattern between forwarding slashes) the forward slash ‘ / ’ has become a special character to be escaped. For example to search for cancer/testis use the query cancer\/testis. If special characters are not escaped the actual query performed may be different from what expected. The OmicsDI Search Box allows the end-users to search data using biological evidence such as the list of the proteins identified in the proteomics experiment or the metabolites reported in the Metabolomics experiment. For example, if the user searches for 3-methyl-2-oxobutanoic in the resource it will find one dataset in Metaboligths and five in Metabolome workbench that identified the current molecule. The final search results are shown in the browser page including Refine Filters.
![]() Institute
Institute
Chongqing University of Posts and Telecommunications (重庆邮电大学)
![]() Author
Author
Yasset Perez-Riverol#, Mingze Bai(白明泽)#, ... Henning Hermjakob #co-first authors
![]() Support
Support
![]() Publication
Publication
![]() Figure
Figure
![]() Funding source
Funding source
[{"id":"2","name":"CNHPP int'l project 2:2014DFB30030(蛋白质组生物大数据及其标准系统)"}]