![]() What's it?
What's it?
dbDEPC (database of Differentially Expressed Proteins in Human Cancer) is designed to store and display differentially expressed proteins (DEPs) in human cancers detected by mass spectrometry (MS) approach. The current version contains 4029 DEPs in 20 cancers, curated from 331 MS experiments in 219 peer-reviewed publications. 56% analysis focused on cancer vs. normal comparison, while others (9%, 11% and 24%) focused on cancer vs. cancer, metastasis study, treatment comparison, respectively. Proteins can be queried through the web-interface using protein name, accession number or sequence similarity, and also browsed by interested cancers or MS experiments. We provided multiple conditions to help you filter the various experiments down to your interested results. The retrieved protein pages contain basic protein description, DEP profiles across multiple caners in each analysis type, related MS experiments, low throughput validation assays and diverse annotations including protein function, sequence, gene ontology and involved pathway. For a set of proteins and multiple cancers, differential expression profile can be visualized in a heatmap to explore the difference and similarity between cancers. In addition, a protein association network is introduced to facilitate the discovery of DEPs associated with your query proteins. Furthermore, the change of protein expression of DEPs is probably relevant to important structural variation in the human genome, such as single amino acid alterations. So these variation sites were highlighted as especially related to development of human cancers. We try to make dbDEPC a continuingly growing resource to facilitate cancer proteomic research and contribute to biomarker discovery.
![]() How to use it?
How to use it?
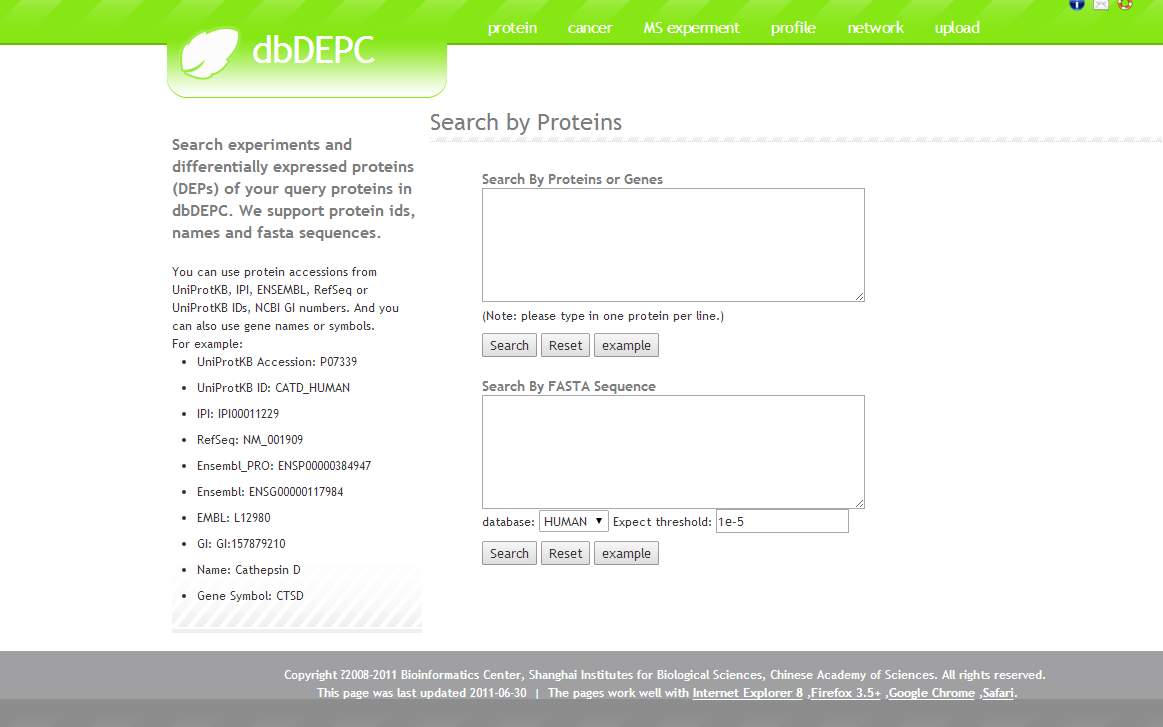
You can search by proteins, cancers or browser all the MS experiments.Then you’ll see related MS experiment results of your queries, and you can use the conditions to filter the results.Later, by clicking the ‘view protein’, you’ll see all the differentially expressed proteins of your queries.This page delineates the detail information for each differentially expressed protein. Basic information is given in the Protein Summary section, including UniportKB Accession, ID, protein description, function, subcellular location, etc.Cancer Profile section reveals the differential expression pattern across multiple cancers in four types of experimental design (Normal vs. Cancer; Metastasis; Treatment; Cancer vs. Cancer).Curated literatures and Experiments details are given in MS Experiment section.Validation Assays section provides other low throughput validation confirmation for the MS indentified differentially expressed protein.Sequence variations are highlighted on the protein sequence. The yellow-marked sites were reported related with cancers, while the green ones come from dbSNP. In Association DEPs section, we provide hyperlinks to other DEPs, which are associated with the DEP on this page with highest confidence score.Annotation information is given in the Function Annotation section, including terms in Gene Ontology and pathway in KEGG.
![]() Institute
Institute
Shanghai Center for Bioinformation Technology (上海生物信息技术研究中心)
![]() Author
Author
Lu Xie(谢鹭)
![]() Support
Support
![]() Figure
Figure
![]() Funding source
Funding source
[{"id":"3","name":"CNHPP int'l project 3:2014DFB30020(中国人类蛋白质组学数据的知识发现)"}]