![]() What's it?
What's it?
GPS-SUMO is a new tool for the prediction of both sumoylation sites and SUMO-interaction motifs (SIMs) in proteins. To obtain an accurate performance, a new generation group-based prediction system (GPS) algorithm integrated with Particle Swarm Optimization approach was applied. By critical evaluation and comparison, GPS-SUMO was demonstrated to be substantially superior against other existing tools and methods. With the help of GPS-SUMO, it is now possible to further investigate the relationship between sumoylation and SUMO interaction processes.
![]() How to use it?
How to use it?
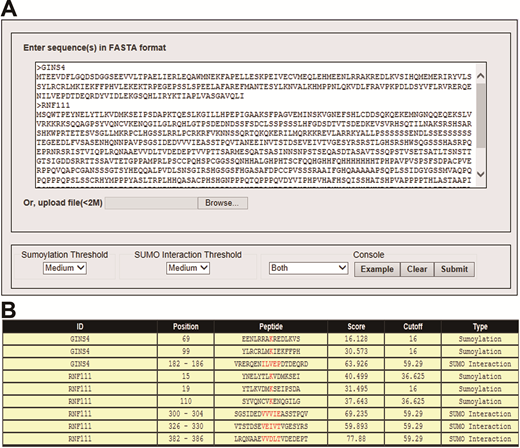
Please visit GPS-SUMO at http://sumosp.biocuckoo.org/down.php. Unlike previous version, the web service does not require a JRE runtime environment. For Windows and Unix/Linux users, please use the keyboard shortcuts "Ctrl+C & Ctrl+V" to copy and paste your FASTA format sequences into TEXT form for prediction. And for Mac users, please use the keyboard shortcuts "Command+C & Command+V". Then please click on the "Submit" button to run the program. The prediction results will be shown in the Prediction form. Again, please click on the ‘Download’ button on the top of the Prediction form to save the results in text or html format.
![]() Institute
Institute
Sun Yat-sen University (中山大学)
![]() Author
Author
Qi Zhao(赵齐), Yu Xue(薛宇), Jian Ren(任间)
![]() Support
Support
![]() Publication
Publication
![]() Figure
Figure
![]() Funding source
Funding source
[{"id":"1","name":"CNHPP int'l project 3:2014DFB30020(中国人类蛋白质组学数据的知识发现)"}]