![]() What's it?
What's it?
Circadian clock exists endogenously in almost every organism and drives oscillatory changes in a myriad of behavioral and physiological processes. Depending on the organism and cell type, the circadian clock promotes rhythmic expression of 1% to over 60% of the genome, serving as the molecular basis for rhythmic control at the system’s level (Li et al., 2015). The CGDB (Circadian Gene DataBase) is an online resource containing over 72,800 genes in more than 148 organisms that exhibit (or may exhibit) daily oscillation at the transcript level. These include 1,382 genes with oscillatory transcripts that have been experimentally validated by techniques including RT-PCR, Northern blot, and in situ hybridization. Because the same gene often exhibits different pattern of temporal expression in different tissues/cells of an organism, and within an organism each tissue/cell type has a unique set of genes that show oscillation at the transcript level (Zhang et al., 2014), we have indicated the phase (time of peak and trough expression) of the oscillation for each gene and the tissue/cell type in which it was identified to be cycling. We have also calculated the amplitude of the oscillation by dividing the peak value with the trough value. Ortholog search for these genes identified another 44,836 which are included in the database as potentially oscillating genes. In addition, we have incorporated 26,582 cycling genes identified in transcriptome profiling studies using microarray or RNA-sequencing. Since post-translational modifications (PTMs) play a key role in regulating circadian timing (Diernfelner et al., 2011; Hardin et al., 2011; Kusakina et al., 2012; Lowrey et al., 2011), we have integrated known PTM sites from published databases including dbPAF (Ullah et al., 2016), dbPPT (Cheng et al., 2014), and CPLM (Liu et al., 2014). All in all, CGDB can serve as a tool to search for genes with oscillatory expression and identify new cycling genes.
![]() How to use it?
How to use it?
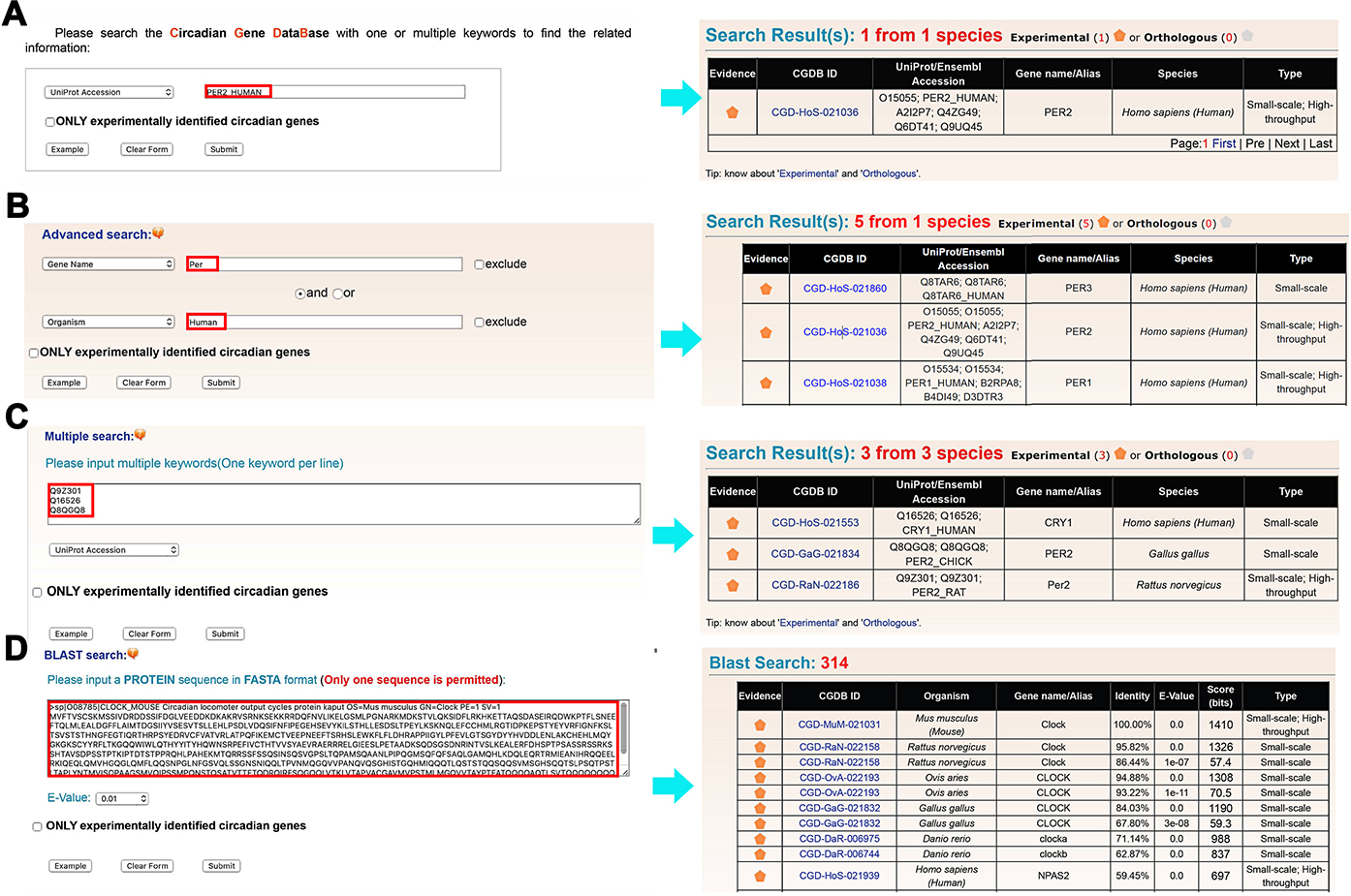
The CGDB database web interface was constructed in an easy-to-use manner. Four search options were provided, including the simple search option providing an interface to query the CGDB database with one or multiple keywords or database accession numbers such as UniProt ID or CGDB ID (Figure 2A), ‘Advanced search’ based on combined keywords with up to two search terms (Figure 2B), ‘Multiple search’ using multiple keywords or accession numbers in a line-by-line format (Figure 2C), and ‘BLAST search’ based on protein sequence (Figure 2D). For example, if a keyword ‘PER2_HUMAN’ in ‘UniProt_Accession’ was submitted for a simple search (Figure 2A), the website will return the circadian gene PER2 from H. sapiens in a tabular format with CGDB ID, UniProt/Ensembl accession, species and gene name/alias (Figure 2A). In advanced search option, two terms specified in two areas are combined with operators ‘and’, ‘or’ and ‘exclude’ to conduct a complex query (Figure 2B). For example, querying the database with ‘Per’ in ‘Gene name’ and ‘Human’ in ‘Organism’ will return five PER genes in H. Sapiens (Figure 2B). Moreover, users could input a list of keywords to perform a multiple search. For example, three core clock genes could be retrieved by querying a list of their UniProt Accessions (Figure 2C). In addition, users could search identical or homologous proteins by submitting a protein sequence in FASTA format in ‘BLAST Search’ (Figure 2D). For example, the FASTA sequence of mouse CLOCK protein could be input in the FASTA format to search for homologous proteins in the database. In particular, there is a checkbox of ‘ONLY experimentally identified circadian genes’ for each search option (Figure 2). Once selected, only experimentally identified cycling genes will be queried and returned.
![]() Institute
Institute
Huazhong University of Science and Technology (华中科技大学)
![]() Author
Author
Ying Zhang(张颖),Yu Xue(薛宇)
![]() Support
Support
![]() Publication
Publication
![]() Figure
Figure
![]() Funding source
Funding source
[{"id":"1","name":"CNHPP int'l project 3:2014DFB30020(中国人类蛋白质组学数据的知识发现)"}]