![]() What's it?
What's it?
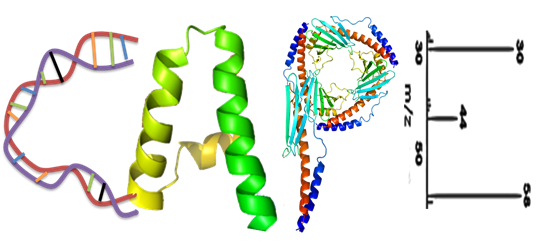
Histone acetylation is a hallmark of chromatin that has an open structure that can be accessed by DNA and RNA polymerases as well as transcription factors, resulting in the activation of gene transcription (Filippakopoulos and Knapp, 2014). Correspondingly, histone methylation increases the basicity and hydrophobicity of histone tails and the affinity of certain proteins, such as transcription factors, toward DNA (Teperino et al., 2010), thus affecting the gene expression. In this database, we have collected 584 non-redundant protein data of 8 organisms including H. sapiens, M. musculus, R. norvegicus, D. melanogaster, C. elegans, A. thaliana, S. pombe and S. cerevisiae from the literature. The data are further classified into 15 families for histone acetylation writers, erasers and readers and 32 families for histone methylation writers, erasers and readers, respectively. WERAM 1.0 is a comprehensive Eukaryotic Writers, Erasers and Readers protein of Histone Acetylation and Methylation system Database for 148 eukaryotic species. And here we provide two approaches for users to browse the database: (i) By species; (ii) By classifications.
![]() How to use it?
How to use it?
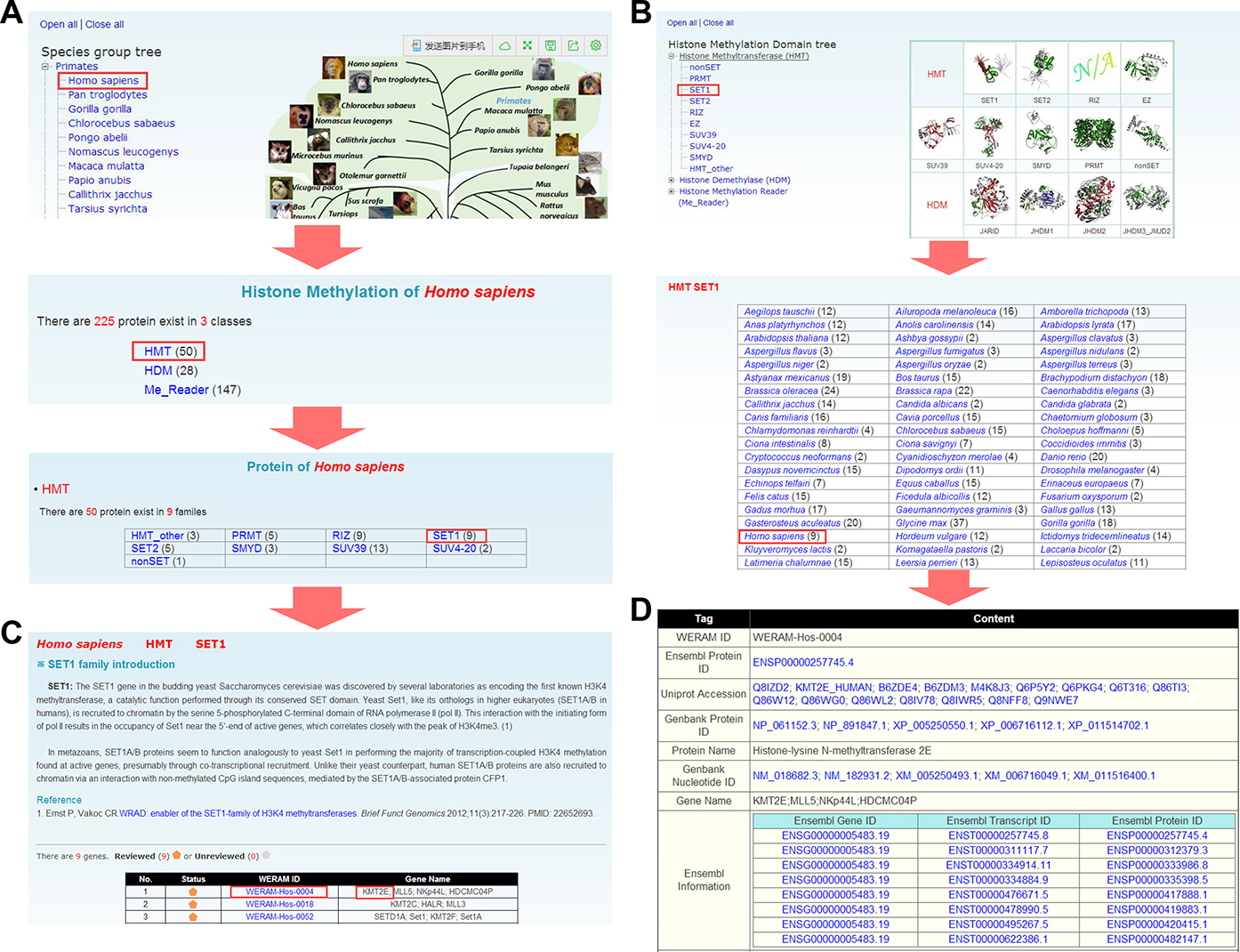
The WERAM database was developed so as to be conveniently used. Here we took a human HMT, Histone-lysine N-methyltransferase 2E (KMT2E), as an example to illustrate the effective usage of WERAM. In order to easily look through the data in WERAM, two approaches were implemented for the browse option: by species or by classification (Figure 2). In the option of ‘Browse by species’, the left tree represents the Ensembl (31) taxonomy categories, including primates, rodents, laurasiatheria and so on, whereas the right tree represents the phylogenetic relationship of the eukaryotic species in Ensembl (Figure 2A). By clicking on the ‘Homo sapiens’ button, the families of HATs, HDACs, HMTs, HDMs and reader proteins in H. sapiens can be visualized (Figure 2A). Since the SET1 family belongs to HMTs, users can click on the ‘HMT’ button to view the family information (Figure 2A). Also, WERAM can be further browsed by classification (Figure 2B). The left tree represents the hierarchical classification, whereas a representative 3D structure of the catalytic or PTM-binding domain was taken from the Pfam database (33) and presented on the right for each family, if available (Figure 2B). Users can click on the ‘SET1’ button to visualize the family information across 148 eukaryotes (Figure 2B). By either clicking on the ‘SET1’ button in the HMT group page (Figure 2A) or the ‘Homo sapiens’ button in the SET1 page (Figure 2B), the members in human SET1 family can be viewed, while a brief description of SET1 functions and regulatory roles is also available (Figure 2C). To organize the database, we used WERAM IDs for the identified HATs, HDACs, HMTs, HDMs and reader proteins, respectively. The Ensembl Gene ID was adopted as the secondary accession (Figure 2C). The users can click on the ‘WERAM-HOS-0004΄ button to view the detailed information of human KMT2E (Figure 2D). In addition, WERAM can be queried in an easy-to-use manner with a number of search and advance options.
![]() Institute
Institute
Huazhong University of Science and Technology (华中科技大学)
![]() Author
Author
Yang Xu(徐阳),Yu Xue(薛宇)
![]() Support
Support
![]() Publication
Publication
![]() Figure
Figure
![]() Funding source
Funding source
[{"id":"1","name":"CNHPP int'l project 3:2014DFB30020(中国人类蛋白质组学数据的知识发现)"}]