![]() What's it?
What's it?
As one of the PTMs with tremendous studies, Phosphorylation regulates a wide variety of biological processes, including signal transduction events (Cohen, 1982). Whereas eukaryotic proteins of phosphorylation have been extensively studied, only limited information is available for phosphorylated proteins in prokaryotic organisms. Previous studies about constructing database of prokaryotic phosphorylation sites mainly focus on tyrosine-, serine, and threonine-phosphorylated proteins (Wurgler-Murphy, King and Kennelly, 2004). However, for the purpose of elucidating the mechanisms of phosphorylation in prokaryotic organisms, other residues which can also be phosphorylated should not be neglected. For example, protein histidine or aspartate phosphorylation plays important roles in two-signal-transduction events (Galperin, Nikolskaya, Koonin, 2001; Swanson, Alex and Simon, 1994). To settle these challenges, we provide a comprehensive database of Prokaryotic Protein Phosphorylation Sites for 7 types of residues, including 7,391 phosphorylation sites in 3,750 proteins.
![]() How to use it?
How to use it?
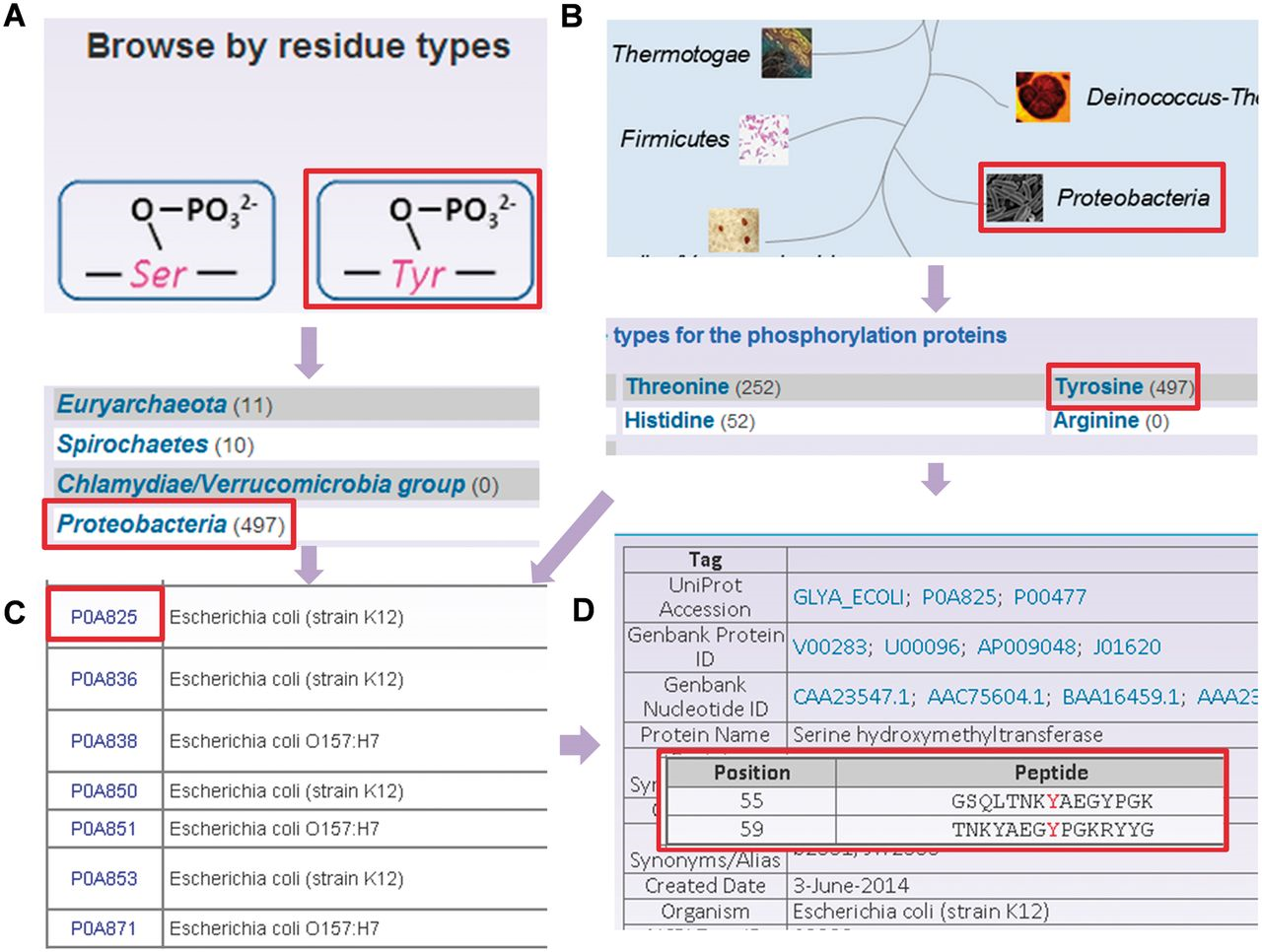
To provide convenient usage, the database was developed in a user-friendly manner, while browse and search options were provided to access the information of prokaryotic phosphorylation sites in the database. Since the phosphorylation sites are identified in different residues and various species, two browse options including ‘Browse by residue types’ ( Figure 3 A) and ‘Browse by phyla’ ( Figure 3 B) were developed in the database. Here, the serine hydroxymethyltransferase in E. coli (strain K12) was selected as an example to describe the usage of browse and search options. In the ‘Browse by residue types’, the phosphorylated residues are shown in diagrams ( Figure 3 A). By clicking the diagram of tyrosine phosphorylation, the distribution of tyrosine phosphorylated phosphoproteins in various organisms is returned ( Figure 3 A). Then the tyrosine phosphorylated phosphoproteins in Proteobacteria could be listed in a tabular format with ‘UniProt Accession’, ‘Name/Alias’ by clicking the link of ‘ Proteobacteria’ ( Figure 3 C). In the option of ‘Browse by phyla’ ( Figure 3 B), the 11 phyla in two domains including bacteria and archaea are listed for users to browse the phosphoproteins ( Figure 3 C). Through clicking on the figure of ‘ Proteobacteria’ , the distribution of phosphoproteins for different modification residue types is shown ( Figure 3 C). Then the list of tyrosine phosphorylated phosphoproteins could be retrieved after clicking the link ‘Tyrosine’, while the detailed information for specific phosphoproteins is provided by clicking protein entry ( Figure 3 D).
![]() Institute
Institute
Huazhong University of Science and Technology (华中科技大学)
![]() Author
Author
Zhicheng Pan(潘智城),Yu Xue(薛宇)
![]() Support
Support
![]() Publication
Publication
![]() Figure
Figure
![]() Funding source
Funding source
[{"id":"1","name":"CNHPP int'l project 3:2014DFB30020(中国人类蛋白质组学数据的知识发现)"}]