![]() What's it?
What's it?
CPLM (Compendium of Protein Lysine Modifications) is an online data resource specifically designed for protein lysine modifications (PLMs). The CPLM database was extended and adapted from our CPLA 1.0 (Compendium of Protein Lysine Acetylation) database (Liu et al., 2011), and the 2.0 release contains 203,972 modification events on 189,919 modified lysines in 45,748 proteins for 12 types of PLMs, including Nε-lysine acetylation (Yang et al., 2007; Shahbazian et al., 2007; Smith et al., 2009), ubiquitination (Gao, et al., 2013), methylation (Chen, et al., 2006), sumoylation (Ren, et al., 2009; Xue, et al., 2006), glycation (Priego-Capote, et al., 2010), butyrylation (Chen, et al., 2007; Cheng, et al., 2009; Zhang, et al., 2009), crotonylation (Tan, et al., 2011), malonylation (Xie, et al., 2012), propionylation (Chen, et al., 2007; Cheng, et al., 2009; Zhang, et al., 2009), succinylation (Xie, et al., 2012; Zhang, et al., 2011), phosphoglycerylation (Moellering, R. E. and B. F. Cravatt, 2013) and prokaryotic Pupylation (Liu, et al., 2011).
![]() How to use it?
How to use it?
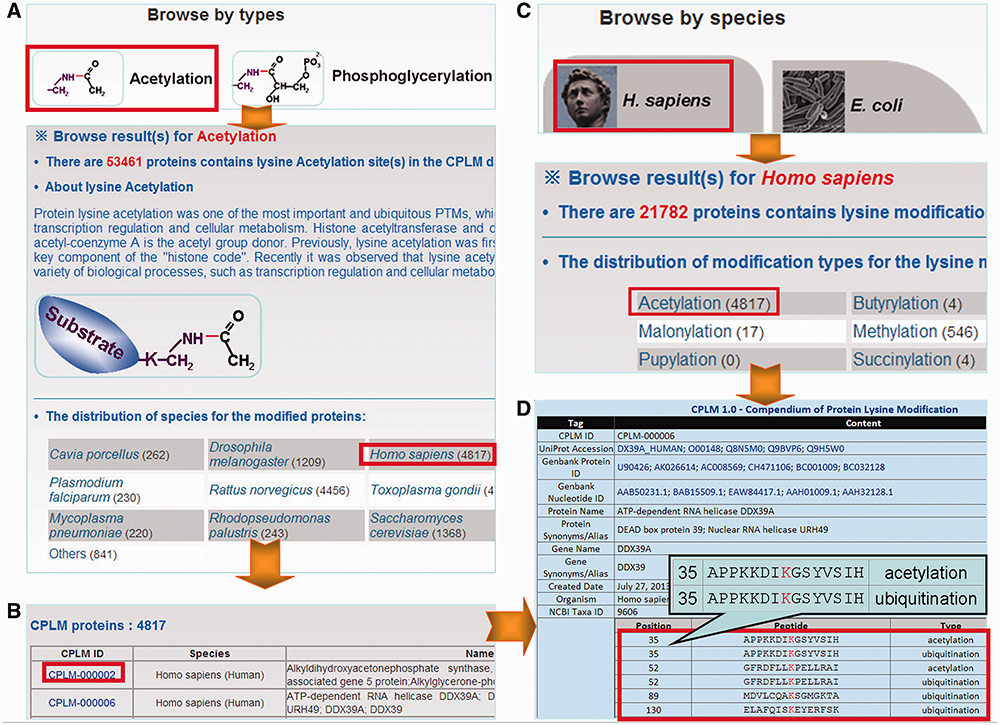
The CPLM database was developed in a user-friendly manner, while browse and search options were provided for accessing the information. Because the proteins and sites could be classified according to the PLM types and species, two browse options including ‘Browse by types’ and ‘Browse by species’ were developed in the database (Figure 2). For convenience, only 12 major species were listed for browsing, while all the other organisms were denoted as ‘Others’. Here, we use lysine acetylation substrates from Homo sapiens as an example to present the usage of the browse options in CPLM. In the option of ‘Browse by types’, 12 simplified molecular structures of ligands conjugated to lysine residues during modification were employed to represent the 12 types of PLMs (Figure 2A). By clicking on the ‘Acetylation’ button, a brief introduction of protein lysine acetylation and the protein number distribution of acetylated proteins in 12 major organisms and other species were showed (Figure 2A). Then the acetylation substrates in H. sapiens could be listed through clicking on the ‘Homo sapiens’ link (Figure 2B). In the option of ‘Browse by species’, the 12 major organisms were organized as animals, bacteria, fungi and plants. Users could click on the ‘H. sapiens’ button to view the protein number distribution of different PLM substrates in H. sapiens (Figure 2C), and then click on the link of ‘Acetylation’ to view the list of acetylated substrates in H. sapiens (Figure 2B). The detailed information for any specified protein could be accessed through the links in the list (Figure 2D).
![]() Institute
Institute
Huazhong University of Science and Technology (华中科技大学)
![]() Author
Author
Zexian Liu(刘泽先),Yu Xue(薛宇)
![]() Support
Support
![]() Publication
Publication
![]() Figure
Figure
![]() Funding source
Funding source
[{"id":"1","name":"CNHPP int'l project 3:2014DFB30020(中国人类蛋白质组学数据的知识发现)"}]