![]() What's it?
What's it?
During cell cycle, numerous proteins temporally and spatially localized in distinct sub-cellular regions including centrosome (spindle pole in budding yeast), kinetochore/centromere, cleavage furrow/midbody (related or homolog structures in plants and budding yeast called as phragmoplast and bud neck, respectively), telomere and spindle spatially and temporally. These sub-cellular regions play important roles in various biological processes. In this work, we have collected all proteins identified to be localized on kinetochore, centrosome, midbody, telomere and spindle from two fungi (S. cerevisiae and S. pombe) and five animals, including C. elegans, D. melanogaster, X. laevis, M. musculus and H. sapiens based on the rationale of "Seeing is believing" (Bloom K et al., 2005). Through ortholog searches, the proteins potentially localized at these sub-cellular regions were detected in 144 eukaryotes. Then the integrated and searchable database MiCroKiTS - Midbody, Centrosome, Kinetochore, Telomere and Spindle has been established. Currently, the MiCroKiTS 4.0 database was updated on Sep. 6, 2014, containing 87,983 unique protein entries. The database will be updated routinely as new microkits proteins are reported.
![]() How to use it?
How to use it?
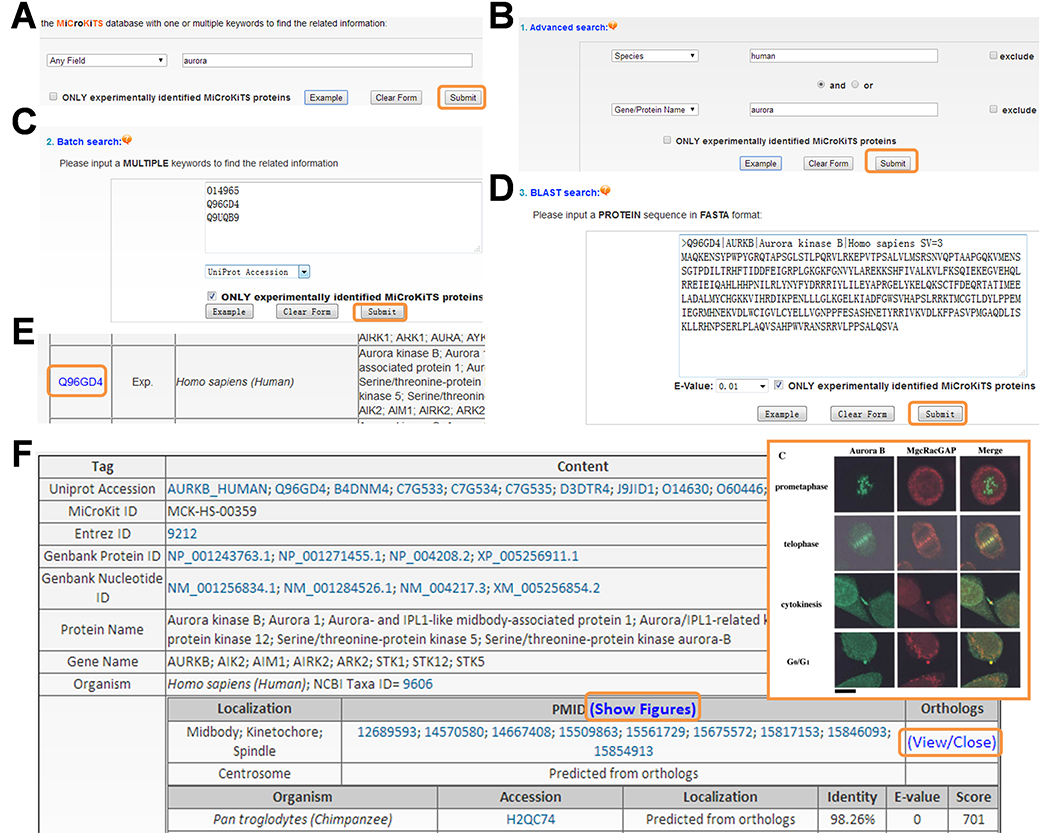
To provide convenient usage, the MiCroKiTS 4.0 database web interface was designed in a user-friendly manner for search and browse. The website contains four search options including one/multiple keywords-based simple search, ‘Advanced search’ based on a combination of multiple keywords, multiple keywords based ‘Batch search’ and protein sequence-based ‘BLAST search’. For example, if a keyword ‘aurora’ in ‘Any Field’ was submitted for a simple search , the website will return a list of MiCroKiTS proteins, such as Aurora kinase B from H. sapiens in a tabular format with accession, species, and protein/gene names/aliases . By clicking the accession ‘Q96GD4’, user could visit the webpage of human Aurora kinase with detailed annotation including localizations, PubMed IDs of source references and the orthologs . For human MiCroKiTS proteins, the representing fluorescent microscope figures were provided by clicking the ‘Show Figures’ . Furthermore, two terms specified in two areas and combined with operators of ‘and’, ‘or’ and ‘exclude’ could be employed to perform a complex query in ‘Advanced Search’ . For example, querying the database with ‘human’ in ‘Species’ and ‘aurora’ in ‘Gene/Protein Name’ will return three human aurora kinases . Alternatively, user could submit a list of keywords to perform a batch search. For example, three human aurora kinases could be retrieved by submitting the list of their UniProt accessions . Furthermore, user could submit a protein sequence in FASTA format in ‘BLAST Search’ to find homologous MiCroKiTS proteins . For example, the sequence of human Aurora kinase B could be input in the FASTA format to search homologous proteins in the database. The ‘Advanced Search’, ‘Batch search’ and ‘BLAST Search’ will return the list of searching hits in a tabular format as the simple search. In addition, there is a checkbox of ‘ONLY experimentally identified MiCroKiTS proteins’ for each search option. If the checkbox is selected, only experimentally identified MiCroKiTS proteins will be queried.
![]() Institute
Institute
Huazhong University of Science and Technology (华中科技大学)
![]() Author
Author
Lili Ma(马丽丽),Yu Xue(薛宇)
![]() Support
Support
![]() Publication
Publication
![]() Figure
Figure
![]() Funding source
Funding source
[{"id":"1","name":"CNHPP int'l project 3:2014DFB30020(中国人类蛋白质组学数据的知识发现)"}]