![]() What's it?
What's it?
The dbPAF (database of Phospho-sites in Animals and Fungi) is an online data resource specifically designed for protein phosphorylation in seven eukaryotic species, including H. sapiens, M. musculus, R. norvegicus, D. melanogaster, C. elegans, S. pombe and S. cerevisiae. From the scientific literature, we collected 294,370 non-redundant phosphorylation sites of 40,432 proteins. We also integrated known phosphorylation sites from a number of public databases, such as Phospho.ELM (Diella, et al., 2004; Diella, et al., 2008), dbPTM (Huang, et al., 2015; Lee, et al., 2006), PHOSIDA (Gnad, et al., 2011; Olsen, et al., 2006), PhosphositePlus (Hornbeck, et al., 2004; Hornbeck, et al., 2015), PhosphoPep (Bodenmiller, et al., 2008; Bodenmiller, et al., 2007), PhosphoGRID (Sadowski, et al., 2013; Stark, et al., 2010), SysPTM (Li, et al., 2009; Li, et al., 2014), HPRD (Goel, et al., 2012) and UniProt (The UniProt Consortium, 2015). In total, dbPAF 1.0 contained 483,001 known phosphorylation sites of 54,148 protein substrates, as a comprehensive data resource for human, animals and fungi.
![]() How to use it?
How to use it?
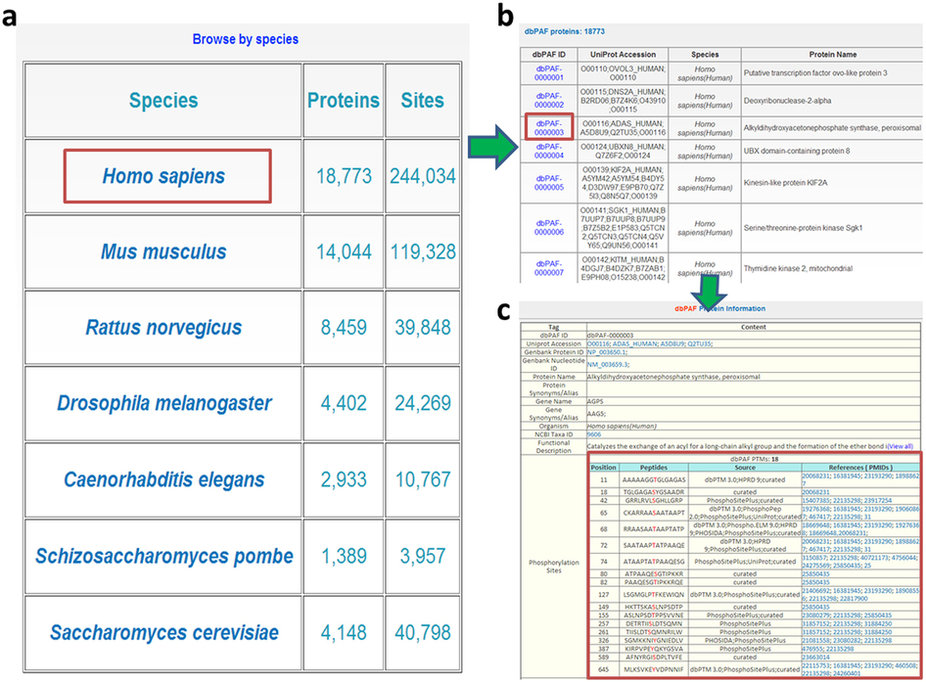
Here we chose human peroxisomal alkyldihydroxyacetonephosphate synthase (AGPS) as an example to demonstrate the usage of “Browse by species”. After clicking on the species diagram of H. sapiens , all human phosphorylated proteins would be listed in a tabular format with “dbPAF ID”, “UniProt Accession”, “Species”, and “Protein Name” (Fig. 3b). A unique “dbPAF ID” was assigned to each protein for the convenient organization of dbPAF database. Then by clicking on the “dbPAF-0000003”, the detailed annotations including 18 known phospho-sites of human AGPS could be shown.
![]() Institute
Institute
Huazhong University of Science and Technology (华中科技大学)
![]() Author
Author
Shaofeng Lin(林少峰),Yu Xue(薛宇)
![]() Support
Support
![]() Publication
Publication
![]() Figure
Figure
![]() Funding source
Funding source
[{"id":"1","name":"CNHPP int'l project 3:2014DFB30020(中国人类蛋白质组学数据的知识发现)"}]