![]() What's it?
What's it?
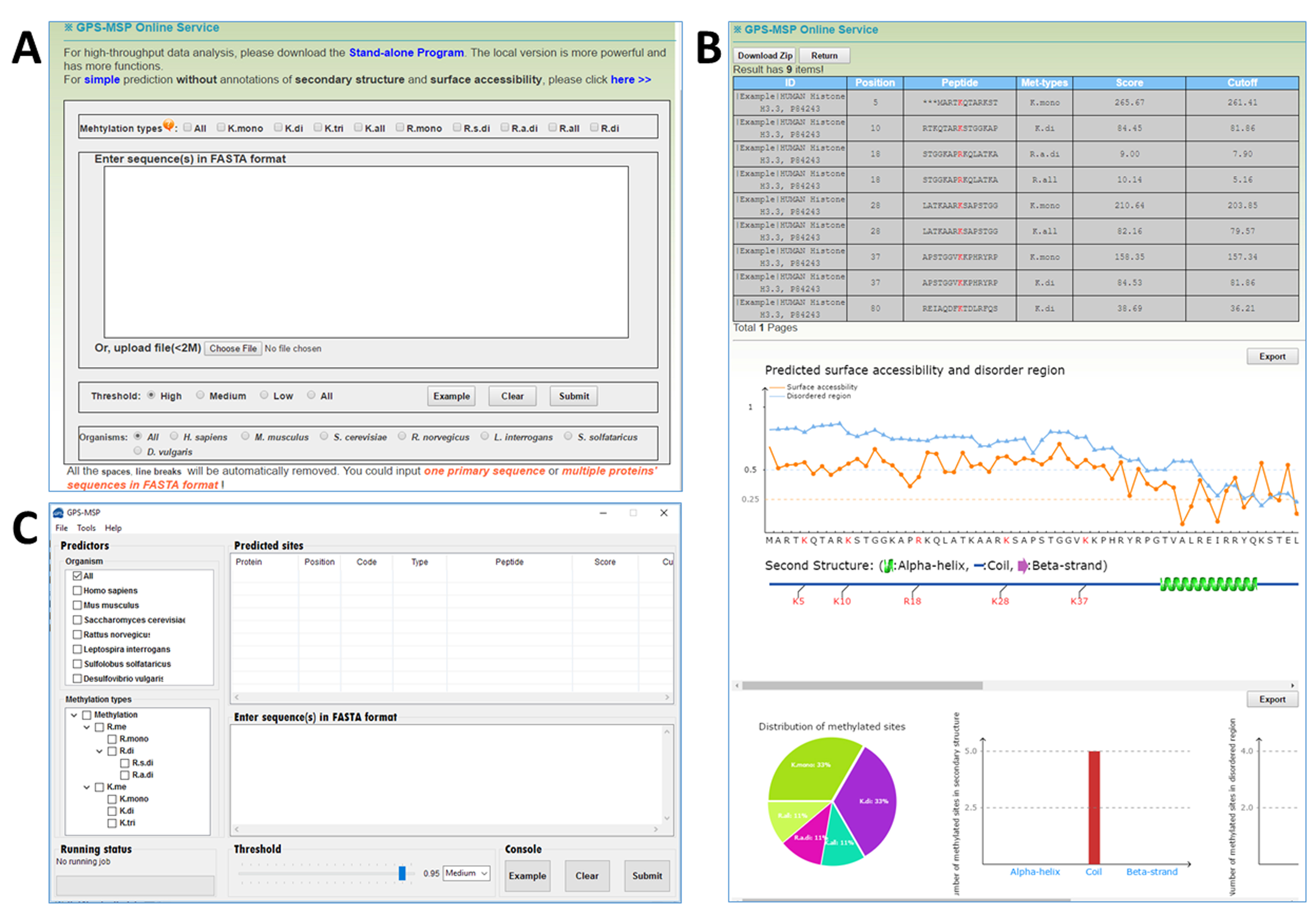
In this work, we adopted GPS 3.0 algorithm and built GPS-MSP (Methyl-group Specific Predictor) for the prediction of general or type-specific methylline and methylarginine residues in proteins. We also construced up to 28 organism-specific predictors. We implemented GPS-MSP into a webserver, which also provides the information of secondary structures, protein surface accessibility, and the statistics of prediction results .
![]() How to use it?
How to use it?
The input of GPS-MSP including 3 major parts: methylation types, protein sequence(s) and threshold. Additionally, users can choose a species-specific predictor for species specific prediction.
![]() Institute
Institute
Huazhong University of Science and Technology (华中科技大学)
![]() Author
Author
Wankun Deng(邓万锟),Yu Xue(薛宇)
![]() Support
Support
![]() Publication
Publication
![]() Figure
Figure
![]() Funding source
Funding source
[{"id":"1","name":"CNHPP int'l project 3:2014DFB30020(中国人类蛋白质组学数据的知识发现)"}]