![]() What's it?
What's it?
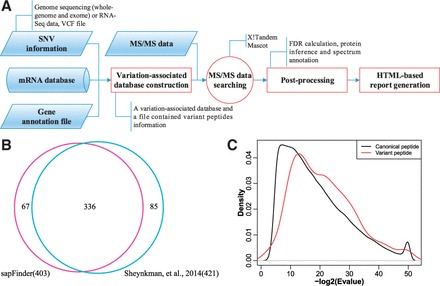
sapFinder, an R software package, for detection of the variant peptides based on tandem mass spectrometry (MS/MS)-based proteomics data. This package automates the construction of variation-associated databases from public SNV repositories or sample-specific next-generation sequencing (NGS) data and the identification of SAPs through database searching, post-processing and generation of HTML-based report with visualized interface.
![]() How to use it?
How to use it?
sapFinder is implemented as a Bioconductor package in R. The package and the document/example can be viewed and downloaded at http://bioconductor.org/packages/devel/bioc/html/sapFinder.html and are provided under a GPL-2 license.
![]() Institute
Institute
Shenzhen Huada Gene Research Institute (深圳华大基因研究院)
![]() Author
Author
Bo Wen(闻博), Shaohang Xu(许少行)
![]() Support
Support
![]() Publication
Publication
![]() Figure
Figure
![]() Funding source
Funding source
[{"id":"1","name":"CNHPP int'l project 3:2014DFB30020(中国人类蛋白质组学数据的知识发现)"}]