![]() What's it?
What's it?
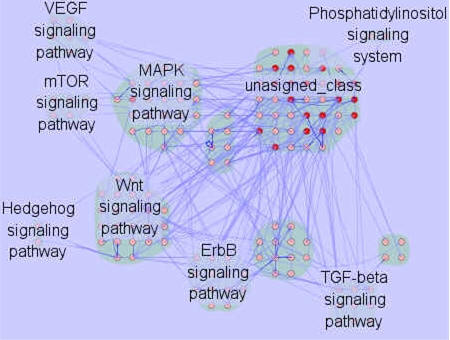
PNmerger (biological Pathway and protein Netwrok merge) is a java based plug-in for the widely used open-source Cytoscape molecular interaction viewer. For an interaction network, PNmerger will automatically annotate the network proteins with the KEGG pathway information, to find the known pathway elements in protein network, and also to predict the possible pathway elements. To present the pathway information for the protein network , PNmerger can illustrate the clusters of the nodes with the same biological pathway, and also present the the potential crosstalks between different pathways. This information will be helpful for the users to find the important clues for knowledge discovery and also experimental design.
![]() How to use it?
How to use it?
When Cytoscape has started, select PNmerger from the Cytoscape plugins menu to show the control panel of PNmerger. There are two options on PNmerger control panel.Step 1-select the organism of the input interaction network. Now, the version 1.0 of PNmerger supports Homo sapiens(human) , Saccharomyces cerevisiae , Caenorhabditis elegans(nematode) , Drosophila melanogaster(fruit fly), Mus musculus(mouse) and Rattus norvegicus(rat).Step 2-select the pathway category: signal transduction pathways/ metabolism pathways/ regulatory pathways. Step 3, click Analyze button. With analyze result, PNmerger add pathway information as an attribute of each node in the input network. If certain node in the input network appears in more than one pathway, PNmerger add several copies of the node to the input network. If certain node appears in k pathways, PNmerger add k-1 copies of that node, each copies is the same with the original node except the pathway attribute. For instance, node A appears in pathway X, pathway Y and pathway Z, PNmerger set pathway X as the pathway attribute, then add two nodes A(copy1) and A(copy2) with pathway Y and pathway Z as their pathway attribute to the input network. If certain node in the input network does not appear in any pathway, PNmerger set unsigned_class as the pathway attribute of it. Also PNmerger add crosstalk information as a node attribute to identify weather certain node is a crosstalk molecule. Integrated another Cytoscape plugin PNmerger, PNmerger can layout network according to the pathway attribute of each nodes. PNmerger can also show crosstalk nodes as an eye-catching color different with other nodes.
![]() Institute
Institute
Beijing Proteome Research Center(BPRC) (北京蛋白质组研究中心)
![]() Author
Author
Dong Li(李栋),Fuchu He (贺福初)
![]() Support
Support
![]() Publication
Publication
![]() Figure
Figure
![]() Funding source
Funding source
[{"id":"1","name":"CNHPP int'l project 3:2014DFB30020(中国人类蛋白质组学数据的知识发现)"}]