![]() What's it?
What's it?
2019nCoVR features comprehensive integration of genomic and proteomic sequences as well as their metadata information from the GISAID, NCBI, NMDC and CNCB/NGDC. It also incorporates a wide range of relevant information including scientific literatures, news, and popular articles for science dissemination, and provides visualization functionalities for genome variation analysis results based on all collected SARS-CoV-2 strains.
![]() How to use
it?
How to use
it?
Enter the strain number of the virus to query its metadata such as lineage, sequence quality and submission time. The functions listed by Popular Resources facilitate users to browse the information of virus epidemic trend, lineage, mutation, clinical diagnosis, etc
![]() Institute
Institute
BEIJING INSTITUTE OF GENOMICS (中国科学院北京基因组研究所)
![]() Author
Author
Shuhui Song(宋述慧), Lina Ma(马利娜), Dong Zou(邹东), Dongmei Tian (田东梅), Cuiping Li (李翠萍), Junwei Zhu (朱军伟)
![]() Support
Support
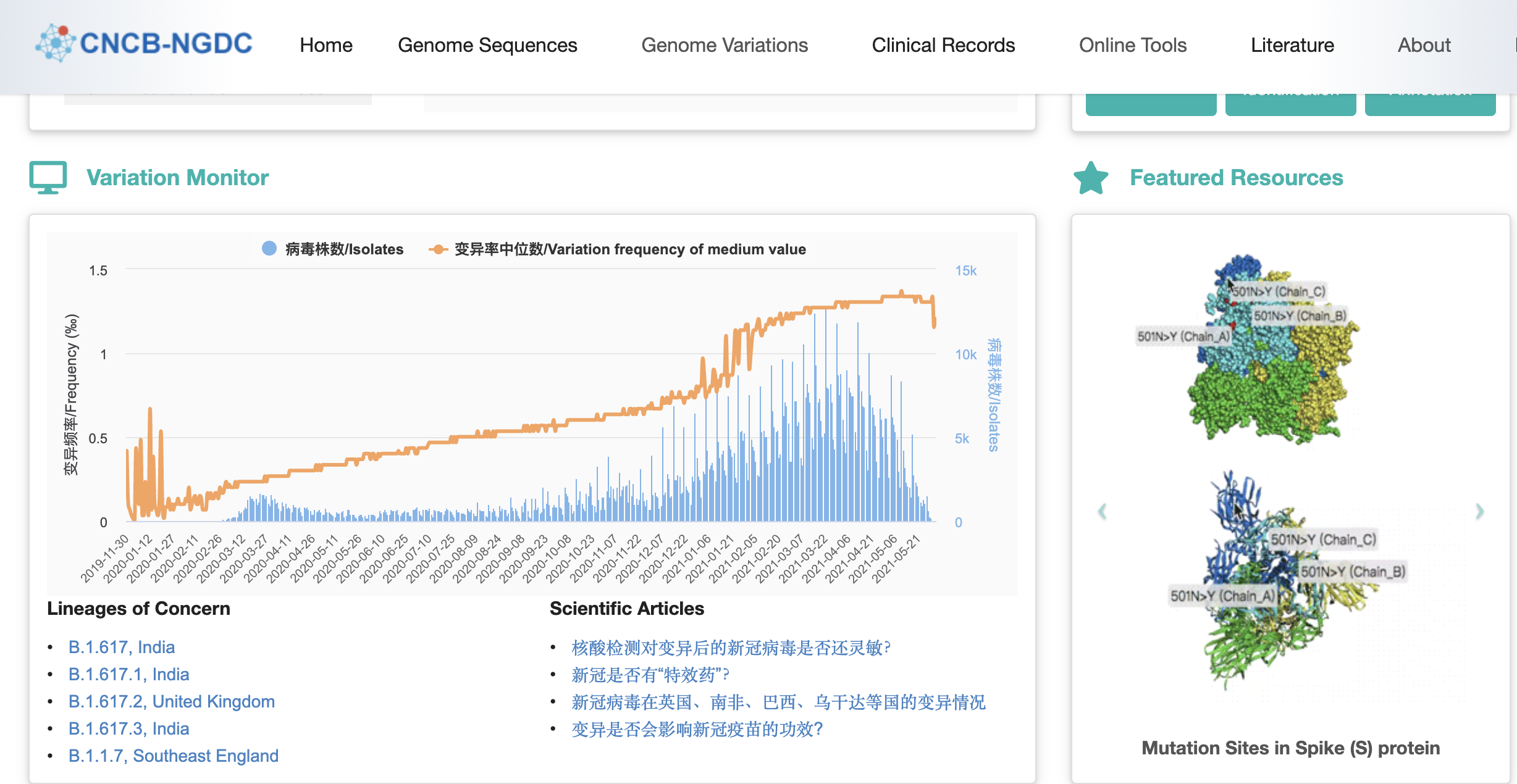
![]() Figure
Figure
![]() Funding
source
Funding
source
[{"id":"China National Center for Bioinformation (CNCB) / National Genomics Data Center (NGDC), and supported by the 13th Five-year Informatization Plan of Chinese Academy of Sciences (No. XXH13505-05), the Strategic Priority Research Program of the Chinese Academy of Sciences (XDA19090116,XDA08020102), the Genomics Data Center Construction of Chinese Academy of Sciences (0202), the Youth Innovation Promotion Association of Chinese Academy of Sciences and CAS Key Technology Talent Program.","name":"China National Center for Bioinformation (CNCB) / National Genomics Data Center (NGDC), and supported by the 13th Five-year Informatization Plan of Chinese Academy of Sciences (No. XXH13505-05), the Strategic Priority Research Program of the Chinese Academy of Sciences (XDA19090116,XDA08020102), the Genomics Data Center Construction of Chinese Academy of Sciences (0202), the Youth Innovation Promotion Association of Chinese Academy of Sciences and CAS Key Technology Talent Program."}]